Abstract
INTRODUCTION Molecular diagnostics have emerged as a powerful tool to diagnose, sub-classify, and assign a more precise prognosis in myeloid neoplasia. In MDS, a condition with a tremendous pathomorphologic and genomic diversity, molecular markers have the potential to improve the precision of the current prognostic schemes, traditionally relying on clinical parameters. The recent IPSSM incorporated the presence of certain somatic mutations as consequential for disease outcomes, including overall (OS) and leukemia-free survival (LFS)1. As 70% of patients used to develop the IPSSM were untreated, which is not reflective of real-world approaches, validation in other cohorts is crucial. Indeed, the increased precision score over the traditional IPSSR was most obvious when a fraction of untreated patients was analyzed separately vs. a modest improvement in treated cases.
METHODS We analyzed a large cohort of MDS patients diagnosed at our institution to validate the novel IPSSM and to compare its prognostication precision vs. actuarial survival with the previously well-established IPSSR. To present a real-world clinical experience, we used a cohort of treated and untreated MDS patients. The same IPSSM algorithm used in the original paper was applied except for the inclusion of the uncommon (<2%) partial tandem duplication of KMT2A (MLL).
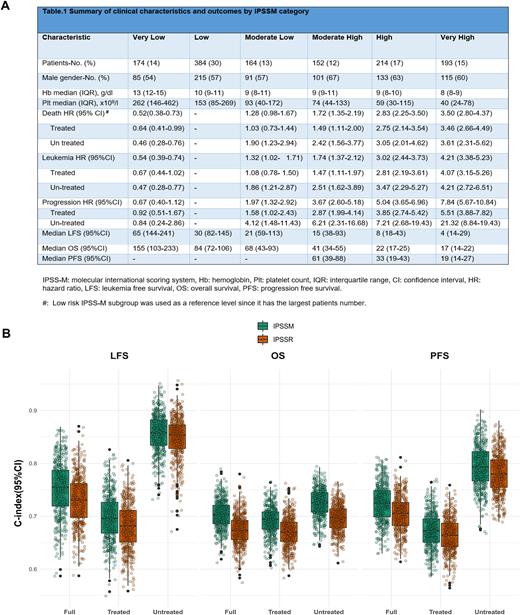
RESULTS Among 1281 MDS patients, the overall, median age at MDS onset was 70 years (IQR: 62-77) with a 1.5 M:F ratio. The median follow-up time was 2.4 years (0.9-5.4). MDS with single lineage dysplasia (SLD) and multilineage dysplasia (MLD) were the most common subtypes (both 38%), followed by MDS with excess blast (EB) in 22% of cases. In this real-life cohort, 60% of the patients received disease modifying treatment ([DMT] (of which 40% consisted of HMAs) vs. 30% in the original IPSSM cohort. As reflected by the IPSSM scores, most patients with complex karyotype (75%), TP53MT (75%), RUNX1MT (64%), and NRASMT (65%) were assigned to High and Very High risk groups with the majority of carriers of biallelic TP53MT(61%) falling in the latter. After applying the IPSSM algorithm, patients were redistributed in Very Low (13%), Low (30%), Moderate Low (13%). Moderate High (12%), High (17%) and Very High (15%) risk categories, (Fig.1, panel A), thus resulting in re-stratification of 45% of the whole patients, similar to the IPSSM publication. Of these, 70% were up-staged and 30% were down-staged. The highest rate of re-stratification was observed among High (59%) IPSSR patients.
To explore the differences between IPSSM and IPSSR in predicting outcomes, 3 endpoints (OS, LFS, and progression free survival [PFS]) were used. Overall, the 3-year OS rate was 51% (95%CI:47.6-53.8) during the follow-up period of 2.4 years (IQE:0.9-5.4). The 3-year LFS and PFS rates were 47% (44.0-50.1) and 65% (61.2-67.6), respectively. Overall, significant differences in OS, LFS, and PFS were observed across the risk categories assigned by IPSSM. However, in a subgroup analysis, IPSSM did not add prognostic value for all the 3 above-mentioned outcomes in patients assigned to High and Very High IPSSR risk subgroups (76% treated). When we applied a reverse analysis, IPSSR failed to re-stratify almost any of the IPSSM subgroups for all 3 endpoint outcomes.
To further confirm our results, we estimated the Harrell's c-index (concordance index) for the full cohort, treated patients, and untreated patients (Fig.1, panel B). For OS, we measured a 2.8 point increase in the c-index from IPSSR to IPSS-M. However, both IPSSR and IPSSM showed similar C-indices for OS, LFS, and PFS. In the untreated cohort, both IPSSM and IPSSR showed improvement in the c-indices, probably due to few 'events’ in the low-risk groups.
CONCLUSION We validated the IPSSM in a real-world MDS setting of treated patients. IPSSM included somatic mutations frequently and non-frequently encountered in MDS, with the latter being important in cases of rare, yet significant molecular lesions and interactions. While IPSSM may identifying higher- or lower-risk patients within individual IPSSR subgroups, it may not add significant prognostic value in real-life scenarios that involve patients receiving DMTs during the course of their disease.
REFERENCES 1. Bernard et al. Molecular International Prognostic Scoring System for Myelodysplastic Syndromes. NEJM Evidence June 12, 2022;1.
Disclosures
Carraway:CTI Biopharma: Consultancy, Membership on an entity's Board of Directors or advisory committees; Stemline: Speakers Bureau; Gilead: Consultancy, Membership on an entity's Board of Directors or advisory committees; Novartis: Honoraria, Speakers Bureau; Takeda: Other: DSMB; Syndax: Other: DSMB; AbbVie: Other: DSMB; Jazz: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; BMS: Consultancy, Honoraria, Speakers Bureau. Sekeres:Novartis: Membership on an entity's Board of Directors or advisory committees; Takeda/Millenium: Membership on an entity's Board of Directors or advisory committees; Bristol Myers-Squibb: Membership on an entity's Board of Directors or advisory committees; Kurome: Membership on an entity's Board of Directors or advisory committees. Maciejewski:Alexion: Consultancy; Apellis Pharmaceuticals: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal